Enhanced Sensitivity of Idelalisib and Ibrutinib-Resistant Cell Lines to Anti-CD38 Antibodies
Article Information
Matera Eva-Laure1, Fouret Julien2,3, Baulu Estelle1, Perrial Emeline1, Bousfiha Zineb1, Abdelkamel Chettab1, Lars Petter Jordheim1, Charles Dumontet1,4*
1Cancer Research Center of Lyon, INSERM 1052/CNRS 5286, University of Lyon, Hospices Civils de Lyon, Lyon, France
2ViroScan3D SAS, Trévoux, France
3CIRI, Centre International de Recherche en Infectiologie, Inserm, U1111, Université Claude Bernard Lyon 1, CNRS UMR5308, ENS de Lyon, Université Lyon, Hospices Civils de Lyon, Lyon, France
4ProfileXpert SFR-Santé Lyon Est, UCBL UMS 3453 CNRS-US7 INSERM, Lyon, France
*Corresponding Author: Dr. Charles Dumontet, Cancer Research Center of Lyon, INSERM 1052/CNRS 5286, University of Lyon, Hospices Civils de Lyon, Lyon, France
Received: 16 January 2020; Accepted: 04 February 2020; Published: 10 February 2020
Citation: Matera Eva-Laure, Fouret Julien, Baulu Estelle, Perrial Emeline, Bousfiha Zineb, Abdelkamel Chettab, Lars Petter Jordheim, Charles Dumontet. Enhanced Sensitivity of Idelalisib and Ibrutinib-Resistant Cell Lines to Anti-CD38 Antibodies. Journal of Cancer Science and Clinical Therapeutics 4 (2020): 071-077.
View / Download Pdf Share at FacebookAbstract
BCR pathway inhibitors idelalisib and ibrutinib were the first small molecule targeted agents for B-cell malignancies. In spite of encouraging response rates in various forms of B cell diseases, patients will eventually develop relapse due to the emergence of resistant cells. To better identify the possible mechanisms of resistance we developed and characterized idelalisib- and ibrutinib-resistant variants of the human non Hodgkin’s lymphoma cell lines DoHH2 and Daudi. These resistant variants displayed a cross-resistance profile limited to PI3K inhibitors, BTK inhibitors and a SYK inhibitor but not to unrelated agents. A number of alterations were observed in the resistant lines, including a strong reduction of BLINK and the AKT3. A new SNP in PLCgamma2 (Leu848Phe) closely related to a previously reported SNP associated to ibrutinib resistance (Leu845Phe) was observed in both DAUDI-based resistant cell lines but was absent in wild type cells. Resistant lines tended to express larger amounts of CD38 and were found to display enhanced sensitivity to anti-CD38 antibodies. These results identify potential novel mechanisms of resistance to idelalisib and ibrutinib and raise the possibility that cells resistant to BCR pathway inhibitors might possess enhanced sensitivity to anti-CD38 antibodies.
Keywords
<p>Inhibitors; Resistant lines; Lymphoma; Anti-CD38 antibodies</p>
Inhibitors articles, Resistant lines articles, Lymphoma articles, Anti-CD38 antibodies articles
Inhibitors articles Inhibitors Research articles Inhibitors review articles Inhibitors PubMed articles Inhibitors PubMed Central articles Inhibitors 2023 articles Inhibitors 2024 articles Inhibitors Scopus articles Inhibitors impact factor journals Inhibitors Scopus journals Inhibitors PubMed journals Inhibitors medical journals Inhibitors free journals Inhibitors best journals Inhibitors top journals Inhibitors free medical journals Inhibitors famous journals Inhibitors Google Scholar indexed journals Resistant lines articles Resistant lines Research articles Resistant lines review articles Resistant lines PubMed articles Resistant lines PubMed Central articles Resistant lines 2023 articles Resistant lines 2024 articles Resistant lines Scopus articles Resistant lines impact factor journals Resistant lines Scopus journals Resistant lines PubMed journals Resistant lines medical journals Resistant lines free journals Resistant lines best journals Resistant lines top journals Resistant lines free medical journals Resistant lines famous journals Resistant lines Google Scholar indexed journals Lymphoma articles Lymphoma Research articles Lymphoma review articles Lymphoma PubMed articles Lymphoma PubMed Central articles Lymphoma 2023 articles Lymphoma 2024 articles Lymphoma Scopus articles Lymphoma impact factor journals Lymphoma Scopus journals Lymphoma PubMed journals Lymphoma medical journals Lymphoma free journals Lymphoma best journals Lymphoma top journals Lymphoma free medical journals Lymphoma famous journals Lymphoma Google Scholar indexed journals Anti-CD38 antibodies articles Anti-CD38 antibodies Research articles Anti-CD38 antibodies review articles Anti-CD38 antibodies PubMed articles Anti-CD38 antibodies PubMed Central articles Anti-CD38 antibodies 2023 articles Anti-CD38 antibodies 2024 articles Anti-CD38 antibodies Scopus articles Anti-CD38 antibodies impact factor journals Anti-CD38 antibodies Scopus journals Anti-CD38 antibodies PubMed journals Anti-CD38 antibodies medical journals Anti-CD38 antibodies free journals Anti-CD38 antibodies best journals Anti-CD38 antibodies top journals Anti-CD38 antibodies free medical journals Anti-CD38 antibodies famous journals Anti-CD38 antibodies Google Scholar indexed journals idelalisib articles idelalisib Research articles idelalisib review articles idelalisib PubMed articles idelalisib PubMed Central articles idelalisib 2023 articles idelalisib 2024 articles idelalisib Scopus articles idelalisib impact factor journals idelalisib Scopus journals idelalisib PubMed journals idelalisib medical journals idelalisib free journals idelalisib best journals idelalisib top journals idelalisib free medical journals idelalisib famous journals idelalisib Google Scholar indexed journals ibrutinib articles ibrutinib Research articles ibrutinib review articles ibrutinib PubMed articles ibrutinib PubMed Central articles ibrutinib 2023 articles ibrutinib 2024 articles ibrutinib Scopus articles ibrutinib impact factor journals ibrutinib Scopus journals ibrutinib PubMed journals ibrutinib medical journals ibrutinib free journals ibrutinib best journals ibrutinib top journals ibrutinib free medical journals ibrutinib famous journals ibrutinib Google Scholar indexed journals malignancies articles malignancies Research articles malignancies review articles malignancies PubMed articles malignancies PubMed Central articles malignancies 2023 articles malignancies 2024 articles malignancies Scopus articles malignancies impact factor journals malignancies Scopus journals malignancies PubMed journals malignancies medical journals malignancies free journals malignancies best journals malignancies top journals malignancies free medical journals malignancies famous journals malignancies Google Scholar indexed journals lymphoproliferative diseases articles lymphoproliferative diseases Research articles lymphoproliferative diseases review articles lymphoproliferative diseases PubMed articles lymphoproliferative diseases PubMed Central articles lymphoproliferative diseases 2023 articles lymphoproliferative diseases 2024 articles lymphoproliferative diseases Scopus articles lymphoproliferative diseases impact factor journals lymphoproliferative diseases Scopus journals lymphoproliferative diseases PubMed journals lymphoproliferative diseases medical journals lymphoproliferative diseases free journals lymphoproliferative diseases best journals lymphoproliferative diseases top journals lymphoproliferative diseases free medical journals lymphoproliferative diseases famous journals lymphoproliferative diseases Google Scholar indexed journals lymphoid malignancies articles lymphoid malignancies Research articles lymphoid malignancies review articles lymphoid malignancies PubMed articles lymphoid malignancies PubMed Central articles lymphoid malignancies 2023 articles lymphoid malignancies 2024 articles lymphoid malignancies Scopus articles lymphoid malignancies impact factor journals lymphoid malignancies Scopus journals lymphoid malignancies PubMed journals lymphoid malignancies medical journals lymphoid malignancies free journals lymphoid malignancies best journals lymphoid malignancies top journals lymphoid malignancies free medical journals lymphoid malignancies famous journals lymphoid malignancies Google Scholar indexed journals chronic lymphocytic leukemia articles chronic lymphocytic leukemia Research articles chronic lymphocytic leukemia review articles chronic lymphocytic leukemia PubMed articles chronic lymphocytic leukemia PubMed Central articles chronic lymphocytic leukemia 2023 articles chronic lymphocytic leukemia 2024 articles chronic lymphocytic leukemia Scopus articles chronic lymphocytic leukemia impact factor journals chronic lymphocytic leukemia Scopus journals chronic lymphocytic leukemia PubMed journals chronic lymphocytic leukemia medical journals chronic lymphocytic leukemia free journals chronic lymphocytic leukemia best journals chronic lymphocytic leukemia top journals chronic lymphocytic leukemia free medical journals chronic lymphocytic leukemia famous journals chronic lymphocytic leukemia Google Scholar indexed journals
Article Details
1. Introduction
The introduction of small molecule kinase inhibitors has deeply modified the treatment of patients with lymphoproliferative diseases [1]. Idelalisib, an oral reversible phosphoinositide-3 kinase (PI3K) p110δ inhibitor and ibrutinib, an oral irreversible inhibitor of Bruton’s Tyrosine Kinase (BTK) are both approved in certain indications of lymphoid malignancies [2]. Despite the initial efficacy of these drugs in a majority of cases, some patients relapse during treatment or fail to respond to single agent therapy, prompting the development of combination regimens [3, 4]. It is therefore necessary to better understand mechanisms of resistance to these agents [5].
Resistance to small molecule targeted therapies is commonly due to alterations in the target molecule. Resistance mechanisms for the Bruton's tyrosine kinase inhibitor ibrutinib have been reported to involve mutations of the binding site of ibrubinib to BTK, potential gain of function mutations of PLCgamma2 and activation of MYC [6, 7]. In the case of chronic lymphocytic leukemia (CLL) patients treated with ibrutinib acquired mutations of BTK or PLCG2 were found in up to 85% (95% CI, 71% to 94%) of CLL patients who relapsed under therapy [8]. SWI-SNF mutations have been shown to mediate resistance to ibrutinib and venetoclax in mantle cells lymphoma [9]. Resistance to idelalisib has been linked to PIK3 and Myc amplification [10].
2. Material and Methods
2.1 Cell lines and cytotoxicity assaysCell lines used were the Burkitt lymphoma line DAUDI, and the follicular lymphoma line DOHH2 (both obtained from ATCC). They were made resistant to idelalisib (=idela-R) or ibrutinib (=ibru-R) by exposing the cells to increasing drug concentrations over several months. In the case of idelalisib Daudi was exposed from 1.3 µM up to a final concentration of 16 µM while DoHH2 was exposed from 2.4 µM up to 15 µM. For ibrutinib resistant variants Daudi was exposed from 0.13 µM to a final concentration of 4 µM while DoHH2 was exposed from 0.04 µM up to 0.6 µM. The half maximal inhibitory concentration (IC50) of several drugs was obtained using MTT assay, as previously described [11].
2.2 RT-PCR
Total RNA was extracted from cell suspensions and 2 µg of mRNA was reverse transcribed. PCR was performed on LC480 LightCycler® Nano in triplicate. Data were calculated using the ?Ct method with ribosomal 28S RNA as endogenous control.
2.3 Western blotting
Proteins were extracted using a RIPA lysis buffer. Antibodies used were anti-total AKT (Cell Signaling, ref: 9272S), anti-AKT1 (Cell signaling, ref: 2967L), anti-AKT3 (Cell Signaling 14983), anti-β actin (Sigma, ref: A5441). Secondary antibodies were anti-rabbit (Eurobio, ref: 926-68071) and anti-mouse (Eurobio, ref: 926-32210).
2.4 Flow cytometry analysisCD38 expression was performed using anti CD38 APC antibody (CD38-APC-HB7, BD, ref: 345807) or APC Mouse IgG1, for isotype control on a FACSCalibur flow cytometer. Apoptosis was evaluated using Annexin V/apoptosis detection kit.
2.5 RNA seq analysis
Poly-A enriched and directional mRNA libraries were prepared from total RNA (NEXTflex Rapid Directional mRNA-Seq Kit, BiooScientific). Pair-end reads were generated (Illumina NextSeq 500) generating 75bp pair-end read per sample. Reads were mapped against human genome (hg19) using tophat2 (v2.1.0). For expression analysis FPKM (Fragment Per Kilobase Million) values were computed at the transcript level using cufflinks (v2.1.1) based on Refseq annotation and differential expression was considered with at least a 2-fold-change. SNP variants were called using samtools mpileup and bcftools. The defined BCR pathway from KEGG database was used to focus more specifically on SNP in genes involved in BCR pathway. SNPs on those genes were manually analysed using VarSome platform (varso.me) if changes between WT and treated cell lines were based on at high quality coverage (base quality above Q30).
3. Results and Discussion
3.1 Resistant variants are specifically resistant to BCR pathway inhibitors
Resistance to therapy is usually multifactorial. Yahiaoui analysed the mechanisms of acquired resistance to BCR inhibitors in the TMD8 line and reported a loss of phosphatase and tensin homolog associated with an enhanced phosphoinositide 3-kinase pathway upregulation, but no mutation of PIK3CD while acquired resistance to Bruton's tyrosine kinase inhibitor was associated with a novel tumor necrosis factor alpha induced protein 3 mutation (TNFAIP3 Q143*), increased p-IκBα and a mutation in Bruton's tyrosine kinase at C481F [12]. Cao et al. reported that the WHIM-like CXCR4(S338X) somatic mutation activates AKT and ERK, and promotes resistance to ibrutinib [13]. Iyengar et al. have shown that P110α-mediated constitutive PI3K signaling limits the efficacy of p110δ-selective inhibition in mantle cell lymphoma [14]. Kuo et al. [15] reported point mutations within the kinase PIM1 that reduce sensitivity to ibrutinib in ABC-DLBCL [15].
Remarkably our ibrutinib- and idelalisib-resistant variants displayed high levels of cross-resistance to BCR-pathway inhibitors but not to agents with unrelated mechanisms of action (Figure 1A). Variants were cross-resistant to the two PI3K inhibitors idelalisib and duvelisib, the two BTK inhibitors ibrutinib and tirabrutinib (ONO-4059) and the SYK inhibitor entospletinib but remained sensitive towards conventional cytotoxic agents as well as bortezomib and prednisone. Resistant Daudi variants displayed some level of resistance to navitoclax but not to venetoclax. These results suggest that alterations in the BCR signaling pathway are common events involved in the resistant phenotype of our variants. Alterations observed in all of the resistant models included increased expression of PI3KCG and a strong reduction of protein tyrosine phosphatases PTPN6 and PTPNPRO, protein tyrosine kinase Lyn and NFK-B interactors IKBKB and MALT1 (Figure 1B.) A remarkable alteration in BCR signaling was a significant decrease in the expression of BLNK in all the resistant models. Additionally Akt3 was identified by RNAseq as being strongly down-regulated in all of the resistant variants. This was further confirmed at the protein level (Figure 1C) while total AKT and Akt1 were present at similar contents in sensitive and resistant lines. Akt3 has been reported to suppress Aurora kinase induced apoptosis and conferred resistance to polo-like kinase inhibitors [16, 17]. Conversely Akt3 was found to be associated with slower disease progression in glioblastoma, as opposed to Akt1 and Akt2 [18]. Exposure of Waldenström’s cell lines to the AKT inhibitor MK2206 and ibrutinib was found to be synergistic [19].
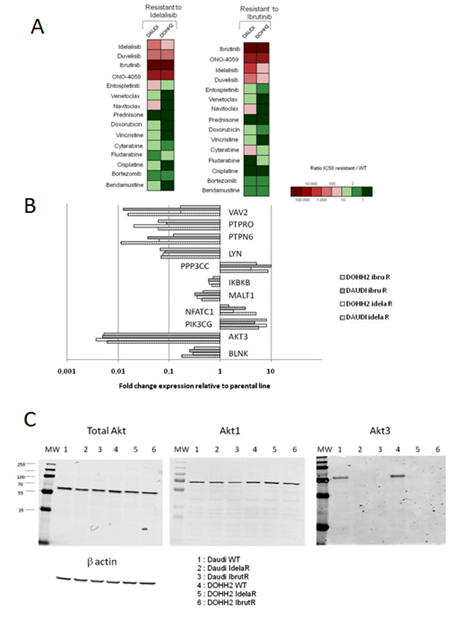
Figure 1: Characteristics of idelalisib- and ibrutinib-resistant cell line variants. A - Sensitivity profiles to BCR pathway inhibitors and unrelated agents; B - expression level alterations; C - Akt3 alterations in resistant variants.
Fifteen SNPs with missense mutations were found on expressed genes involved in the BCR pathway (hsa05220), but no premature stop codon nor frameshift mutation in coding sequences were identified (Supplemental Table 1). SNPs previously reported on BTK and PLCgamma2 were not found in the resistant variants, despite the expression of concerned genes. Of note a new SNP in PLCgamma2 (Leu848Phe) closely related to a previously reported SNP associated to ibrutinib resistance (Leu845Phe) was present on half of expressed transcripts in both DAUDI-based resistant cell lines but was absent in wild type cells. The RNAseq expression analysis identified an upregulation of CD38 in DAUDI and DOHH2 cell lines resistant to idelalisib, a result confirmed by flow cytometry (Figure 2A). Variants were exposed to daratumumab and isatuximab and were found to undergo enhanced apoptosis in comparison to parental cells (Figure 2B). It would be worthwhile to determine whether the enhancement of CD38 is also observed in the tumor microenvironment since CD38 targeting has been proposed as a possible means to reduce local immunosuppression [20].
These in vitro models of resistance to BCR inhibitors suggest novel mechanisms of resistance and have identified enhanced CD38 expression as a possible therapeutic target to be explored and validated in patients with resistant disease.
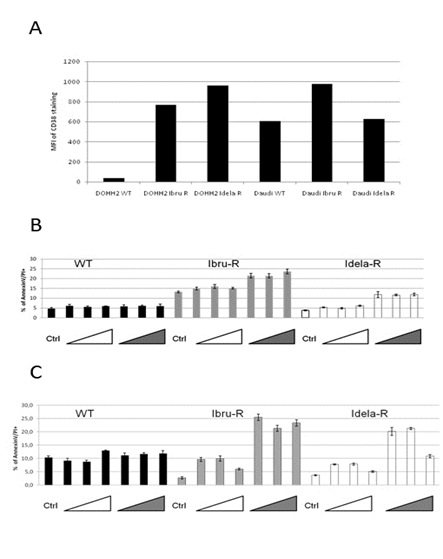
Figure 2: CD38 expression is increased in resistant variants and these are more sensitive to anti CD38 antibodies. A - Expression of CD38 evaluated by flow cytometry in idelalisib- and ibrutinib-resistant variants; B - Sensitivity of parental cell lines and resistant variants to apoptosis induced by anti CD38 antibodies daratumumab (white triangles) and isatuximab (grey triangles) in vitro.
Acknowledgements
This work was supported in part by the LYRICAN grant (INCa_INSERM_DGOS_12563). The PhD fellowship of JF is co-founded by the DGA (Direction Générale de l’Armement) and Viroscan3D.
Authorship Contributions
ELM, EB, EP, ZB and AC performed experiments. JF performed RNA seq analyses. LPJ and CD designed and interpreted the results. CD wrote the manuscript which was reviewed and validated by all co-authors.
Disclosure of Conflicts of Interest
The authors have no conflicts of interest to disclose.
Significance Statement
There are few data regarding mechanisms of resistance to the BCR-pathway inhibitors ibrutinib and idelalisib. Our results suggest novel mechanisms of resistance to these agents and a potential reversal strategy using anti CD38-targeted antibodies.
References
- Jerkeman M, Hallek M, Dreyling M, et al. Targeting of B-cell receptor signalling in B-cell malignancies. J Intern Med 282 (2017): 415-428.
- Abou Zahr A, Bose P, Keating MJ. Pharmacotherapy of relapsed/refractory chronic lymphocytic leukemia. Expert Opin Pharmacother 18 (2017).
- Jerkeman M, Eskelund CW, Hutchings M, et al. Ibrutinib, lenalidomide, and rituximab in relapsed or refractory mantle cell lymphoma (PHILEMON): a multicentre, open-label, single-arm, phase 2 trial. Lancet Haematol 5 (2018): e109-e116.
- Zelenetz AD, Barrientos JC, Brown JR, et al. Idelalisib or placebo in combination with bendamustine and rituximab in patients with relapsed or refractory chronic lymphocytic leukaemia: interim results from a phase 3, randomised, double-blind, placebo-controlled trial. Lancet Oncol 18 (2017): 297-311.
- Lampson BL, Brown JR. Are BTK and PLCG2 mutations necessary and sufficient for ibrutinib resistance in chronic lymphocytic leukemia? Expert Rev Hematol 11 (2018): 185-194.
- Lee J, Zhang LL, Wu W, et al. Activation of MYC, a bona fide client of HSP90, contributes to intrinsic ibrutinib resistance in mantle cell lymphoma. Blood Adv 2 (2018): 2039-2051.
- Woyach JA, Furman RR, Liu TM, et al. Resistance mechanisms for the Bruton's tyrosine kinase inhibitor ibrutinib. N Engl J Med 370 (2014): 2286-2294.
- Woyach JA, Ruppert AS, Guinn D, et al. BTKC481S-Mediated Resistance to Ibrutinib in Chronic Lymphocytic Leukemia. J Clin Oncol 35 (2017): 1437-1443.
- Agarwal R, Chan YC, Tam CS, et al. Dynamic molecular monitoring reveals that SWI-SNF mutations mediate resistance to ibrutinib plus venetoclax in mantle cell lymphoma 25 (2018): 119-129.
- Woyach JA, Johnson AJ. Targeted therapies in CLL: mechanisms of resistance and strategies for management. Blood 126 (2015): 471-477.
- Hage-Sleiman R, Herveau S, Matera EL, et al. Silencing of tubulin binding cofactor C modifies microtubule dynamics and cell cycle distribution and enhances sensitivity to gemcitabine in breast cancer cells. Mol Cancer Ther 10 (2011): 303-312.
- Yahiaoui A, Meadows SA, Sorensen RA, et al. PI3Kdelta inhibitor idelalisib in combination with BTK inhibitor ONO/GS-4059 in diffuse large B cell lymphoma with acquired resistance to PI3Kdelta and BTK inhibitors. PLoS One 12 (2017): e0171221.
- Cao Y, Hunter ZR, Liu X, et al. The WHIM-like CXCR4(S338X) somatic mutation activates AKT and ERK, and promotes resistance to ibrutinib and other agents used in the treatment of Waldenstrom's Macroglobulinemia. Leukemia 29 (2015): 169-176.
- Iyengar S, Clear A, Bodor C, et al. P110alpha-mediated constitutive PI3K signaling limits the efficacy of p110delta-selective inhibition in mantle cell lymphoma, particularly with multiple relapse. Blood 121 (2013): 2274-2284.
- Kuo HP, Ezell SA, Hsieh S, et al. The role of PIM1 in the ibrutinib-resistant ABC subtype of diffuse large B-cell lymphoma. Am J Cancer Res 6 (2016): 2489-2501.
- Noguchi K, Hongama K, Hariki S, et al. Functional Effects of AKT3 on Aurora Kinase Inhibitor-induced Aneuploidy. J Biol Chem 292 (2017): 1910-1924.
- Nonomiya Y, Noguchi K, Tanaka N, et al. Effect of AKT3 expression on MYC- and caspase-8-dependent apoptosis caused by polo-like kinase inhibitors in HCT 116 cells. Cancer Sci 107 (2016): 1877-1887.
- Joy A, Kapoor M, Georges J, et al. The role of AKT isoforms in glioblastoma: AKT3 delays tumor progression. J Neurooncol 130 (2016): 43-52.
- Paulus A, Akhtar S, Yousaf H, et al. Waldenstrom macroglobulinemia cells devoid of BTK(C481S) or CXCR4(WHIM-like) mutations acquire resistance to ibrutinib through upregulation of Bcl-2 and AKT resulting in vulnerability towards venetoclax or MK2206 treatment. Blood Cancer J 7 (2017): e565.
- van de Donk N. Immunomodulatory effects of CD38-targeting antibodies. Immunol Lett 199 (2018): 16-22.
