Antimicrobial Resistance in the Central African Region: A Review
Article Information
Patrick Achiangia Njukeng, Denis Ebot Ako-Arrey*, Elvis Tajoache Amin, Charles Njumkeng, Frankline Sevidzem Wirsiy
Global Health Systems Solutions (GHSS), Denveur layout, Bonamoussadi, Littoral Region, Republic of Cameroon
*Corresponding Author: Dr. Denis Ebot Ako-Arrey, Global Health Systems Solutions (GHSS), Denver-Bonamoussadi, Littoral Region, Cameroon
Received: 17 June 2019; Accepted: 03 July 2019; Published: 15 July 2019
Citation:
Patrick Achiangia Njukeng, Denis Ebot Ako-Arrey, Elvis Tajoache Amin, Charles Njumkeng, Frankline Sevidzem Wirsiy. Antimicrobial Resistance in the Central African Region: A Review. Journal of Environmental Science and Public Health 3 (2019): 358-378.
View / Download Pdf Share at FacebookAbstract
Background: The impact of antimicrobial resistance (AMR) has placed it among one of the top public health problems worldwide. AMR has increased the global infectious disease burden and put a greater strain on health systems. This review sought to contribute more evidence on this issue of AMR in the Central African Region, by determining the overall bacterial pathogens resistance rate and pathogen specific resistance rate with respect to antimicrobial agents common in the region.
Methods: Pubmed and other relevant databases were searched using the Boolean strategy. We reviewed articles from 2008 to 2018 and in accordance with an adapted PRISMA guideline. Article retrieval and screening were done using a structured search inclusion/exclusion criteria. Median and interquartile ranges of percentage resistance were calculated for each antibiotic-bacterium combination.
Results: Limited AMR data was available for Central African countries with no reported data for Burundi. A total of 30 articles were included in the final analysis. The most commonly reported bacterium was Salmonella spp that has been reported from 16 studies with median resistance rate of 45.5 (IQR 9.1-81.0). It was also observed that bacterium Staphylococcus aureus had the highest resistance rate with a median resistance of 90 (IQR 86.4-95.2). In general, median resistance rate higher above 50% were observed for the following bacteria; Staphylococcus aureus, Shigella spp, Klebsiella spp, Enterococci, E. Coli and Acinetobacter spp.
Conclusion: The review highlights two important findings: first, there is a huge gap of data regarding Antimicrobial resistance in the Central Afri
Keywords
Antimicrobial resistance (AMR), Central Africa, Bacteria, Review, Resistance rate
Antimicrobial resistance (AMR) articles Antimicrobial resistance (AMR) Research articles Antimicrobial resistance (AMR) review articles Antimicrobial resistance (AMR) PubMed articles Antimicrobial resistance (AMR) PubMed Central articles Antimicrobial resistance (AMR) 2023 articles Antimicrobial resistance (AMR) 2024 articles Antimicrobial resistance (AMR) Scopus articles Antimicrobial resistance (AMR) impact factor journals Antimicrobial resistance (AMR) Scopus journals Antimicrobial resistance (AMR) PubMed journals Antimicrobial resistance (AMR) medical journals Antimicrobial resistance (AMR) free journals Antimicrobial resistance (AMR) best journals Antimicrobial resistance (AMR) top journals Antimicrobial resistance (AMR) free medical journals Antimicrobial resistance (AMR) famous journals Antimicrobial resistance (AMR) Google Scholar indexed journals Bacteria articles Bacteria Research articles Bacteria review articles Bacteria PubMed articles Bacteria PubMed Central articles Bacteria 2023 articles Bacteria 2024 articles Bacteria Scopus articles Bacteria impact factor journals Bacteria Scopus journals Bacteria PubMed journals Bacteria medical journals Bacteria free journals Bacteria best journals Bacteria top journals Bacteria free medical journals Bacteria famous journals Bacteria Google Scholar indexed journals Resistance rate articles Resistance rate Research articles Resistance rate review articles Resistance rate PubMed articles Resistance rate PubMed Central articles Resistance rate 2023 articles Resistance rate 2024 articles Resistance rate Scopus articles Resistance rate impact factor journals Resistance rate Scopus journals Resistance rate PubMed journals Resistance rate medical journals Resistance rate free journals Resistance rate best journals Resistance rate top journals Resistance rate free medical journals Resistance rate famous journals Resistance rate Google Scholar indexed journals public health articles public health Research articles public health review articles public health PubMed articles public health PubMed Central articles public health 2023 articles public health 2024 articles public health Scopus articles public health impact factor journals public health Scopus journals public health PubMed journals public health medical journals public health free journals public health best journals public health top journals public health free medical journals public health famous journals public health Google Scholar indexed journals AMR articles AMR Research articles AMR review articles AMR PubMed articles AMR PubMed Central articles AMR 2023 articles AMR 2024 articles AMR Scopus articles AMR impact factor journals AMR Scopus journals AMR PubMed journals AMR medical journals AMR free journals AMR best journals AMR top journals AMR free medical journals AMR famous journals AMR Google Scholar indexed journals public health articles public health Research articles public health review articles public health PubMed articles public health PubMed Central articles public health 2023 articles public health 2024 articles public health Scopus articles public health impact factor journals public health Scopus journals public health PubMed journals public health medical journals public health free journals public health best journals public health top journals public health free medical journals public health famous journals public health Google Scholar indexed journals animal health articles animal health Research articles animal health review articles animal health PubMed articles animal health PubMed Central articles animal health 2023 articles animal health 2024 articles animal health Scopus articles animal health impact factor journals animal health Scopus journals animal health PubMed journals animal health medical journals animal health free journals animal health best journals animal health top journals animal health free medical journals animal health famous journals animal health Google Scholar indexed journals antibiotics articles antibiotics Research articles antibiotics review articles antibiotics PubMed articles antibiotics PubMed Central articles antibiotics 2023 articles antibiotics 2024 articles antibiotics Scopus articles antibiotics impact factor journals antibiotics Scopus journals antibiotics PubMed journals antibiotics medical journals antibiotics free journals antibiotics best journals antibiotics top journals antibiotics free medical journals antibiotics famous journals antibiotics Google Scholar indexed journals
Article Details
1. Introduction
The impact of antimicrobial resistance (AMR) has placed it among one of the top public health problems worldwide [1] The emergence of AMR and its rapid acceleration among priority pathogens has hindered the effectiveness of antibiotic therapy in many clinical conditions [2]. This has resulted in increased mortality, morbidity as well as higher socio-economic costs. Projections have shown that at this rate, by 2050, an estimated 10 million deaths will occur annually with an associated cumulative economic cost of about US$100 trillion worldwide [2, 3]. AMR has therefore increased the global infectious disease burden and put a greater strain on health systems. It is critical that every country, regardless of income level, should proactively take steps against AMR. Containment will only occur with concerted, coordinated global action [2]. Inaction breeds the risk of reversing the progress made against public health threats of today, such as HIV and malaria, and a likely resurgence of diseases of the past, including infections that were hitherto once treatable like Pneumonia [4]. The World Health Organization (WHO) has recognized the public health threat of AMR and has continuously urged countries to combat it [2]. A WHO global action plan for AMR was adopted in 2015 with recommendations for countries to develop a national AMR action plan, re-inforce surveillance systems, and provide a knowledge and evidence base for action and advocacy, in order to enhance the collection, and analyses of standardized, comparable, and validated data on AMR that can be shared among countries [5]. However, a recent status report on the African region showed that only 2 countries (4.3%) had a national AMR plans in place while only 7 countries (14.9%) have overarching national infection prevention and control (IPC) policies [6]. Additionally, in Africa, significant gaps in surveillance, standard methodologies and data sharing have been reported elsewhere [7].
In line with this Global Action Plan, and to meet needs specific to Africa, the Africa Centers for Disease Control is establishing Anti-Microbial Resistance Surveillance Network (AMRSNET) which is a network of public health institutions and leaders from human and animal health sectors who will collaborate to measure, prevent, and mitigate harms from AMR organisms [3]. This will be achieved by improving surveillance of AMR organisms among humans and animals, delaying emergence of AMR, limiting transmission of AMR and mitigating harm among patients infected with AMR organisms [3]. Despite this global recognition, in some parts of the world like in Central Africa, the sheer magnitude of AMR threat is not fully understood. Available data shows that the Central African Region shares the worldwide trend of increasing drug resistance [8, 9] with resistance being reported for every major class of antibiotic in both community and health-care settings. The emergence, persistence and transmission of AMR patterns in developing countries have been growing as a result of a myriad of factors; genetic capabilities of pathogens, high burden of microbial infections, lack of monitoring of antimicrobial resistance patterns, indiscriminate use of antibiotics, non-judicious prescription by healthcare providers, poor access to diagnostics, unavailability of second line antibiotics, substandard and/or counterfeit antibiotics, inadequate infection prevention and control strategies, globalization, socio-economic factors and fragile health systems [3, 10, 11].
Even though some studies have reported a high prevalence of AMR in sub-Saharan African countries [7], overall, there is still a scarcity of evidence on the real impact of AMR in the region which is further compounded by a considerable variation in this existing data. A WHO global report on antimicrobial resistance, showed that the African region has one of the largest gaps in available evidence on the prevalence of AMR [5]. Also, an external quality assessment reported considerable deficits in antimicrobial susceptibility testing in many African countries [12]. Some studies in Central Africa have reported on the trends of antimicrobial resistance in specific clinical syndromes, while others have focused on specific bacteria isolates [13]. However, there is still limited and up to date information on AMR representation in the Central African Region. The last review of published literature on bacterial resistance in Central Africa was carried out 10 years ago and revealed alarming resistance rates in nearly all pathogens in the region [13]. A situational analysis of antimicrobial drug resistance in Africa stated that the expansion of resources and the technological advances observed in the past decades has led to the availability of larger amounts of drugs in developing countries than ever before, with antibiotics being the most frequently prescribed drugs in hospitals [14]. The dearth of evidence of AMR in the region can be attributed to a number of factors ranging from the lack of established national or regional AMR surveillance systems, inadequate laboratory capacity, lack of resources, weak infrastructures and limited standard operating procedures [3]. It is in this light that, our review sought to update the evidence on this topic, by determining the overall resistance rate of bacterial pathogens and reported resistance of each pathogen with respect to antimicrobial agents common in the Central African region. Establishing the resistance rates will enhance and provide stakeholders with a current status, identified gaps and modifiable causes of AMR in the region.
2. Methodology
The review team comprised of experts in microbiology, anti-microbial resistance, epidemiology, medicine, and systematic reviews methodology. The research design that was applied to this study was a review of the available studies on anti-microbial resistance in Central African Countries. The review permitted was designed to conduct a comprehensive search for primary studies on the focused research questions, selecting studies using clear and reproducible eligibility criteria, critically appraising study quality and completing a synthesis of findings according to pre-determined and explicit methods [15]. Data from all studies on AMR in Central African states were combined and observed for trends and extend of uniformity in the available results. We it was anticipated that by studying similar outcomes across a wide variety of contexts, settings and study designs, we would be able to assess the rigor of available evidence on AMR in the region. Additionally, considering that reviews are considered among the best source of evidence [16], the study was also intended to provide pooled estimates about the impact of AMR in the region, which may be more reliable than evidence from single studies [17].
2.1 Search strategy
We initially conducted a scoping search done to identify existing reviews on AMR in Central Africa, and this permitted us to further highlight relevant search terms and clarify inclusion and exclusion criteria [18] as well as avoid duplication of efforts. For this review, we searched several electronic databases for published and unpublished articles from 2008 to 2018. We decided to start our search in 2008 to coincide with the last published review on this topic in Central Africa [13]. We used the Boolean strategy to search through the following databases; Pubmed, Medline, Embase, Web of Sciences, Google scholar, Cochrane Library, African Journal Online, MEDLINE, and Scopus. In consultation with regional AMR experts, colleagues and members of our research team, we focused on priority microbial pathogens and associated disease conditions that are common within the Central African Region. We used a combination of Medical Subject Heading (MeSH) and free text terms to search through these databases using the following key words;
Antimicrobial resistance, drug resistance, antibiotics, antifungal, antiviral, antiprotozoal, microbial surveillance, antimicrobial susceptibility, antimicrobial sensitivity, Vibro. Cholerae, Staphylococcus aureus, Methicillin-resistant Staphylococcus aureus, Shigella spp, Salmonella spp, Pseudomonas spp., Mycobacterium Tuberculosis, Klebsiella spp, Enterococci, E. Coli, Acinetobacter spp., Non-typhoid Salmonella, Salmonella enterica serovar Typhi, Central Africa, Economic Community of Central African States (ECCAS), Gabon, Cameroon, Central African Republic (CAR), Chad, Congo Brazzaville Equatorial Guinea, Burundi, Rwanda, the Democratic Republic of Congo (DRC), Sao Tome and Principe.
We sought to find relevant databases for research in the region, but none was identified for this study. In order to further extend our evidence base, and minimize publication bias due to selective availability of papers, we decided to search for grey literature (conference abstracts, research reports, book chapters, unpublished data, dissertations and policy documents). Finally, we manually combed through bibliographies and performed hand searching of key journals on the topic. The most recent comprehensive search for each database was March 25th, 2019, and all relevant studies were exported to Zotero bibliographic software.
2.2 Eligibility criteria
According to our study design and the research topic, we established predefined criteria for study retention in the review as follows
2.2.1 Study content: All published (and in press) research articles focusing on Antimicrobial (Bacterial) Drug Resistance (including MDR and TDR).
2.2.2 Timeframe: Papers are eligible if published in or after the year 2008.
2.2.3 Context: We sought studies carried out in any of the 10 Economic Community of Central African States (ECCAS) Countries which include; Gabon, Cameroon, the Central African Republic (CAR), Chad, Congo Brazzaville and Equatorial Guinea, Burundi, Rwanda, the Democratic Republic of Congo (DRC) and Sao Tome and Principe.
2.2.4 Study design/setting: Cross-sectional, longitudinal, longitudinal, Prospective and surveillance studies were sourced either conducted within a Laboratory, hospital or community environment.
2.2.5 Population: Individuals of all age groups and sex.
2.2.6 Setting: Community, laboratory and healthcare facilities.
2.2.7 Language: English and French
2.3 Studies with the following characteristics were excluded
- Antimicrobial resistance studies that focused on animal subjects
- Antimicrobial resistance studies that focused on antifungal, antiviral and antiprotozoal
- Studies that reported data for less than 10 patients
- Studies that reported data for less than 10 isolates of a particular pathogen
2.4 Data screening and extraction
We first of all removed all duplicate articles that we found from the various databases. We then performed an initial screening of the titles and abstracts on the basis of the eligibility criteria stated above in order to validate their selection as part of this review. Next, we performed full text screening of selected studies. All the articles that met our inclusion criteria were retained for data extraction. This was done using an electronic standardized data extraction template that was designed by the team in line with the study objectives, the inclusion criteria, and made up of relevant study components for data analysis. This data extraction template was first pilot tested on a representative sample of articles. The titles, abstract and full text screening as well as data extraction was done independently and in triplicate (ETA, CN and FSW) with disagreements resolved via consensus, or by a tie breaker (DEAA).
2.5 Data analysis
The data was extracted data into an Excel sheet and exported to SPSS version 21 for analysis. Considering that the studies on AMR in the Central African region were heterogeneous and methodologically diverse (context, population, study design, setting, and type of AMR outcomes), a formal study quality assessment nor a meta-analysis of the selected papers could not be done. The data was collated, summarized and categorized in order to perform appropriate statistical analyses. Median statistics was performed and inter quarter range computed if more than three papers had assessed the resistance rates. We calculated median resistance (MR) and interquartile range (IQR) of resistance for each bacterium antibiotic combination; a standardized measure from the collected data. Also, Meta-analysis was not conducted because of the large variability in AMR methodology, geography and the small number of articles available per country. Since the number of studies from hospital/in-patient settings was small, they were combined and median percentages with interquartile ranges were generated.
3. Results and Discussions
3.1 Data and study characteristics
After a careful examination, taking into consideration the inclusion and exclusion criteria, Out of 130 studies that were scoped, 30 studies were found eligible for the review. The adapted PRISMA flow diagram as shown in Figure 1 presents the inclusion and exclusion process.
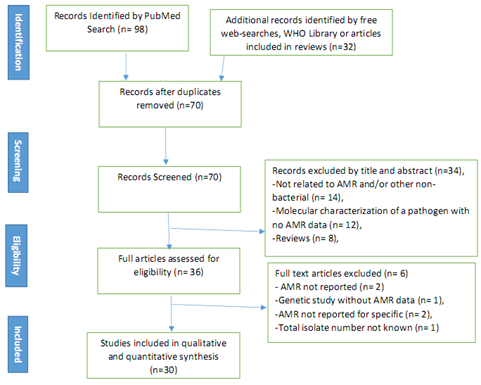
Figure 1: PRISMA Diagram of the article selection procedure for articles published between the year 2008 and 2018, Central Africa.
It worthy to note that, out of the 30 studies that were included for this review, seven of the studies were carried out in Cameroon [10, 19-24], 6 in the Democratic Republic of Congo [25-30], 5 in Gabon [31-35], 4 in Central Africa Republic [36-39] and Rwanda [40-43] and one study each in Chad [44], Congo Brazaville [45], Equatorial Guinea [46], Sao Tome and Principe [47] respectively. We found no study for Burundi. Table 1 presents the number of AMR studies reviewed in the Central Africa Region per member country.
|
Central Africa Region-Member Country |
Number of study reporting antimicrobial activities |
Percentage |
|
Cameroon |
7 |
23.3 |
|
Central Africa Republic |
4 |
13.3 |
|
Chad |
1 |
3.3 |
|
Congo Brazaville |
1 |
3.3 |
|
Democratic Republic of Congo |
6 |
20 |
|
Equatorial Guinea |
1 |
3.3 |
|
Gabon |
5 |
16.7 |
|
Rwanda |
4 |
13.3 |
|
Sao Tome and Principe |
1 |
3.3 |
|
Burundi |
0 |
0 |
|
Total |
30 |
100 |
Table 1: Number of studies review in Central Africa Region per member country (2008-2018).
3.2 Overall bacterial patterns resistance to commonly used antimicrobial drugs
The most commonly reported bacterium was Salmonella spp and been reported by 16 studies with a median resistance rate of 45.5 (9.1-81.0). We also observed that Staphylococcus aureus had the highest median resistance rate of 90 (86.4- 95.2) while the least median resistance rate was observe for Mycobacterium Tuberculosis 5.3 (0.4- 9.3). In general, median resistance rates above 50% were observed for Staphylococcus aureus, Shigella spp, Klebsiella spp, Enterococci, E. Coli and Acinetobacter spp. as shown in Table 2.
|
Pathogen |
Proportion of studies reporting antimicrobial activities to the microbe (%) |
Median of resistance rate MR% (IQR) |
|
Vibro. Cholerae |
1 (3.2) |
44.8 (9.5-97.5) |
|
Staphylococcus aureus |
8 (26.7) |
90 (86.4-95.2) |
|
Methicillin-resistant Staphylococcus aureus |
1 (3.2) |
86.35 (54.5-100) |
|
Shigella spp |
5 (16.1) |
70.5 (31.0-80.0) |
|
Salmonella spp |
16 (51.6) |
45.5 (9.1- 81.0) |
|
Pseudomonas spp. |
5 (16.1) |
18.8 (7.1-81.8) |
|
Mycobacterium Tuberculosis |
2 (6.5) |
5.3 (0.4- 9.3) |
|
Klebsiella spp |
8 (25.8) |
83.4 (58.0-91.7) |
|
Enterococci |
3 (10.0) |
56.5 (21- 100) |
|
E. Coli |
6 (19.4) |
74.1 (33.0-94.0) |
|
Acinetobacter spp. |
2 (6.5) |
80 (2.1-88.0) |
|
Non-typhoid Salmonella |
2 (6.5) |
33.25 (11.6-53.8) |
MR-Median Resistant Rate; IQR-Inter-quartile Range
Table 2: Overall Median Resistance Rate of Pathogens common in Central Africa, (2008-2018).
3.3 Multidrug resistant tuberculosis (MDR TB)
Although overall global incidence is falling, tuberculosis (TB) still claims the lives of approximately 1.6 million people each year [44]. In an attempt to slow the development of resistance, patients are treated with multiple drugs over the course of several months. The long duration of treatment, plus medication side effects and other factors, makes it difficult for patients to adhere to a full course of treatment. As a result, the Directly Observed Treatment, Short Course (DOTS) strategy has been widely adopted to improve adherence. However, an estimated 3% of new TB cases and 20% of previously treated cases have been found to have multidrug resistant TB (MDR-TB). Some estimates indicate that the incidence rate of MDR-TB could double in the next 15 to 20 years; in the next 35 years, it is possible that MDR-TB will claim over 75 million lives. A study by Minime-Lingoupou et al. [19] reported a relatively low primary resistance to anti-tuberculosis drugs in Bangui and Bimbo, Central Africa Republic. In Cameroon, Noeske J. et al. [48] in 2016 reported that fluoroquinolone resistance in MDR-TB patients compromises favourable treatment outcomes. They went ahead to establish an epidemiological and clinical data of patients from Equatorial Guinea put on standardized short-course MDR-TB treatment in Cameroon between (2013-2015) in which 12 of them had an under treatment outcome following failure after first treatment, failure after (several) retreatments and some experiencing relapse after first treatment. This scenario clearly showed that MDR-TB treatment programme can be jeopardised by cross-border migration as seen in Cameroon. Table 3 shows the Median of resistance rate (IQR) of Mycobacterium Tuberculosis to Rifampicin (0.4%), Ethmabutol (2.2%), Streptomycin (8.4%) and Isoniazid (9.3%).
|
Pathogen |
Antibiotic |
Median of resistance rate% (IQR) |
|
Mycobacterium Tuberculosis |
Rifampicin |
0.4 (à) |
|
Ethmabutol |
2.2 (à) |
|
|
Streptomycin |
8.4 (à) |
|
|
Isoniazid |
9.3 (à) |
à = IQR could not be calculated because we had less than three papers that assessed resistance rates.
Table 3: Median Resistance rate of Mycobacterium Tuberculosis to 4 antibiotics in Central Africa, 2008-2018.
In Chad, a study [44] was first; to show that drug resistance of M. tuberculosis was present in the country and established that resistance to isoniazid was the most frequent resistance observed, among 135 patients, with 18 isolates (13%) presenting at least this resistance, Three isolates (2.2%) were resistant to isoniazid and rifampicin (multidrug resistance MDR) including one isolate being concomitantly resistant to streptomycin and ethambutol. A study by Gavin et al. [49] brought to light the potential dissemination of an MDR strain named MDR-TBEG in Equatorial Guinea, a country where little is known about the extent and features of TB or MDR TB. It also highlights that MDR strains can spread across continents, and thus MDR TB’s emergence in any country becomes a global problem. In this study, all 10 patients had an MDR TB rate >2.0% of all combined (new and previously treated) TB cases. In Equatorial Guinea, a study by Tudo, et al. [49] showed that, the overall resistance rate in new cases was 16.9% compared to 41.6% in previously treated cases.
Isoniazid resistance was the most frequent (12.5% and 16.6% respectively) in the two groups (new cases and previously treated cases), while multidrug resistance was observed in 1.7% and 25% of new and previously treated cases, respectively. It has been shown that level of MDR-TB among TB patients in Rwanda is high compared to other Central African states. This was establish that Umubyeyi et al. [50], of 616 strains from new cases, 6.2% were resistant to isoniazid, 3.9% to rifampicin and 3.9% were multidrug-resistant TB. Among 85 strains from previously treated cases, the prevalence of resistance was 10.6%, 10.6% and 9.4% (MDR-TB strains) respectively. Eight MDR cases showed additional resistance to ethambutol and streptomycin. In Cameroon, a study by Noeske et al. [48] reported 26 patients (12%) out of 438 retreatment cases harbored multi-drug resistant (MDR) strains in the Littoral region of the country.
3.4 Extensively drug-resistant tuberculosis (XDR TB)
Worryingly, cases of extensively drug-resistant TB (XDR-TB) have also occurred, although more infrequently. Owing to the fact that XDRTB is resistant to many first-line and reserve TB medicines, the current treatment options are extremely limited. In some cases, these strains of TB are virtually untreatable. However, we didn’t find any published study conducted on extensively drug-resistant TB (XDR-TB) among the 10 Economic Communities of Central African States (ECCAS).
3.5 Vibrio cholerae resistance
Only 1 study [51] reported Vibrio cholerae resistance. In the study, Vibrio cholerae was resistant to Nalidixic acid (18.4%), Erythromycin (9.5%), Cotrimoxazole (97.5%) and Nitrofurantoin (71.1%). This study revealed that Vibrio cholera has a very high (>90%) resistance rate to Cotrimoxazole. Most studies published were describing outbreaks among rural semi-urban dwellers, with V. cholerae O1 El Tor occurring more. A study conducted in DRC showed that 1, 093 Vibrio cholerae isolated from 1997–2012 were found to have increasing antimicrobial drug resistance over time. Further search findings demonstrated that the 2011–2012 epidemic was caused by an El Tor variant clonal complex with a single antimicrobial drug susceptibility profile. However, extensive and multidrug resistance was described from outbreaks in Central African Republic, DRC, with strains resistant to first-line drugs including ampicillin, tetracycline, doxycycline, SXT, nalidixic acid. Chloramphenicol. Table 4 shows the median resistance rates of Vibro. Cholerae to 4 antibiotics.
|
Pathogen |
Antibiotic |
Median of resistance rate% (IQR) |
|
Vibro. Cholerae |
Nalidixic acid |
18.4 (à) |
|
Erythromycin |
9.5 (à) |
|
|
Cotrimoxazole |
97.5 (à) |
|
|
Nitrofurantoin |
71.1 (à) |
à = IQR could not be calculated because we had less than three papers that assessed resistance rates.
Table 4: Median Resistance rate of Vibro. Cholerae to 4 antibiotics in Central Africa, 2008-2018.
3.6 Staphylococcus aureus resistance
In Central African states, resistance data for S. aureus were reported in 8 studies [32, 45, 52-56, 59], including two surveillance studies and six clinical studies from urban hospitals. Although a wide range of samples (pus, wounds, urine and CSF) were examined community or hospital acquired S. aureus infections, only one study considered blood cultures. It is worthy to note, out of this 8 studies, in 3 of them, panels for susceptibility testing were not in accordance with WHO surveillance standards, e.g. did not contain oxacillin or an equivalent antibiotic. In addition, Staphylococcus Aureus median resistant rate to Penicillin was the highest 92.5% (IQR 88.4-98.8) while the least median resistant rate was observed for Gentamycin 1.6% (IQR, 0.9-2.3) as shown on table 5.
|
Pathogen |
Antibiotic |
Median of resistance rate% (IQR) |
|
Staphylococcus Aureus |
Penicillin |
92.5 (88.4-98.8) |
|
Oxacillin |
82 (à) |
|
|
Erythromycin |
47 (6.1- 83.0) |
|
|
Tetracycline |
62 (à) |
|
|
Gentamycin |
1.6 (0.9-2.3) |
|
|
Ciprofloxacin |
54.5 (à) |
à = IQR could not be calculated because we had less than three papers that assessed resistance rates.
Table 5: Median Resistance rate of Staphylococcus Aureus to 6 antibiotics in Central Africa, 2008-2018.
3.7 Methicillin-resistant Staphylococcus aureus (MRSA)
Methicillin-resistant Staphylococcus aureus (MRSA) is a global health concern, but there are few data from Central Africa. Methicillin-resistant Staphylococcus aureus (MRSA) is associated with difficult-to-treat infections and high levels of morbidity [53]. Furthermore, methicillin-resistant Staphylococcus aureus (MRSA) infections are a serious global problem, with considerable impact on patients and substantial health care costs [55]. The epidemiologic changes in MRSA over the years have shown that the distinction between community acquired (CA)-MRSA and hospital-acquired (HA)-MRSA is no longer clear. MRSA data was reported by five studies. Few articles exist in Central African States on the topic of MRSA as they are pertinent to practitioners of manual therapy. Only one study from Cameroon (Gonsu et al.) reported on MRSA. The study analyses the resistance of MRSA to 10 antibiotics, and revealed that MRSA showed 100% resistance 5 (Cefixime, amoxillinclav, amoxicillin, Cefotaxime and Ceftazidime) of the drugs studied while the least 18.2% resistance was observed for cefuroxime. The overall mediate resistance rate of MRSA to the ten antibiotics was 86.35 (IQR 54.5- 100) as showed in table 6.
|
Pathogen |
Antibiotic |
Median resistance rate (IQR) |
|
Methicillin-resistant Staphylococcus aureus (MRSA) |
Ceftazidime |
100 (à) |
|
cefuroxime |
18.2 (à) |
|
|
ceftriaxone |
66.2 (à) |
|
|
Cefotaxime |
100 (à) |
|
|
amoxicillin |
100 (à) |
|
|
Ciprofloxacin |
54.5 (à) |
|
|
amoxillinclav |
100 (à) |
|
|
Norfloxacin |
72.7 (à) |
|
|
Amikacin |
36.4 (à) |
|
|
Cefixime |
100 (à) |
à = IQR could not be calculated because we had less than three papers that assessed resistance rates.
Table 6: Methicillin-resistant Staphylococcus aureus (MRSA) to 10 antibiotics in Central Africa, 2008-2018.
3.8 Shigella spp resistance
Five studies [23, 34, 36, 37, 57] were found in our search. Two-thirds reported on outbreak investigations, and Shigella dysenteriae type 1 was the predominant serotype. The five studies that were found in the search studied the resistance of Shigella Spp to 12 antibiotics and revealed that Shigella showed resistance of over 70% of 6 drugs (amoxicillin, Tetracycline, Trimethroprim sulfamethoxazole, doxycycline and Strptomycin). On the other hand shigella was least resistance to Cefotaxime and gentamycin (10.6% and 0.34% resistance respectively). A study by Njunda et al. [23] in Cameroon, reported that of 223 stool samples cultured, 10 (4.5%) yielded Shigella species. They went ahead to show that Isolation rate was observed to be more in children below 15 years (7.89%), and also higher in rural areas (6.35%). This study also revealed that all 10 isolates showed resistance to at least two antibiotics and 9 (90%) were multi-drug resistant. The highest resistance rates were encountered with tetracycline (98.5%) and streptomycin (97%). Least resistance was observed with gentamycin (034%). Table 7 shows the median resistance rates of Shigella spp to 12 antibiotics.
|
Pathogen |
Antibiotic |
Median of resistance rate% (IQR) |
|
Shigella spp |
Cefotaxime |
10.6 (à) |
|
amoxicillin |
80 (à) |
|
|
amoxillinclav |
58.5 (à) |
|
|
ciprofloxacine |
66 (à) |
|
|
Nalidixicacid |
47.0 (à) |
|
|
gentamycin |
0.34 (à) |
|
|
Tetracycline |
98.5 (à) |
|
|
Trimethroprim sulfamethoxazole |
86 (à) |
|
|
chloramphenicol |
69.5 (à) |
|
|
Doxycycline |
83 (à) |
|
|
Streptomycin |
97 (à) |
|
|
kanamycin |
55 (à) |
à = IQR could not be calculated because we had less than three papers that assessed resistance rates.
Table 7: Median Resistance rate of Shigella spp. to 12 antibiotics in Central Africa, 2008-2018.
3.9 Salmonella spp. resistance
It is worthy no note that, Salmonella enterica serotype Typhi and the nontyphoid S. enterica (NTS) are leading causes of bacteremia in Africa, though little information is available from Central Africa. Table 8 shows the median resistance rates of Salmonella spp to antibiotics. Sixteen studies [25-29, 37, 38, 43, 55-53, 58-62] from 2008 onwards addressed S. Typhi, mainly in urban settings in Cameroon, Equatorial Guinea, DRC and Gabon. Five of them were surveillance studies, of which they were conducted in DRC. Isolates were recovered from blood and faeces samples. Although median resistance rates to amoxicillin, chloramphenicol and SXT were relatively low, there was a steeply increasing upward trend towards 50% resistance rates. Combined resistance was described in at least four studies. In a Multidrug-Resistant Salmonella enterica, study in Democratic Republic of the Congo, Phoba et al. [25] found resistance to four or more first-line antibiotics in >95% of the isolates. Table 8 shows the median resistance rates of Salmonella spp to 23 antibiotics.
|
Pathogen |
Antibiotic |
Median of resistance rate% (IQR) |
|
Salmonella spp |
Cefotaxime |
2.2 (à) |
|
Amoxicillin |
82 (à) |
|
|
Amoxillinclav |
38.5 (à) |
|
|
Ofloxacin |
20 (à) |
|
|
Ciprofloxacine |
35.5 (à) |
|
|
Nalidixicacid |
7 (3.6-50.3) |
|
|
Gentamycin |
5 (0.4-6.0) |
|
|
Tetracycline |
72.8 (34.8-92.6) |
|
|
Trimethroprimsulfamethoxazole |
80.8 (62.4-92.6) |
|
|
Chloramphenicol |
68.0 (35.0-88.1) |
|
|
Azithromycin |
3.3 (0.8-11.4) |
|
|
Doxycycline |
83 (à) |
|
|
Ampicillin |
77.7 (50.2- 92.5) |
|
|
Streptomycin |
27 (à) |
|
|
kanamycin |
55 (à) |
|
|
Cotrimoxazole |
90 (à) |
|
|
Cefalothin |
4.0 (à) |
|
|
Multidrugresistance |
79.7 (35.9-87.5) |
|
|
DecreasedciprofloxacinsusceptibilityDCS |
2.2 (1.8-15.4) |
|
|
Extendedspectrumlactamase |
6.9 (à) |
|
|
Sulphoamide |
29 (à) |
|
|
Ticarcillin |
30 (à) |
|
|
Ampicillinbetalactamaseinhibitor |
3.9 (à) |
à = IQR could not be calculated because we had less than three papers that assessed resistance rates.
Table 8: Median Resistance rate of Salmonella spp. to 23 antibiotics in Central Africa, 2008-2018.
3.10 Non-typhoid Salmonella (NTS) resistance
Two studies [27, 28] specifically addressed Non-typhoid Salmonella (NTS) resistance in the Central African region. The general study characteristics were similar to those reporting on S. Typhi. As part of a microbiological surveillance study in DR Congo, blood cultures were collected between 2007 and 2011. Isolated NTS were assessed for serotype and antimicrobial resistance, including decreased ciprofloxacin susceptibility and extended-spectrum beta-lactamase (ESBL) production. In total, 233 NTS isolates (representing 23.6% of clinically significant organisms) were collected, mainly consisting of Salmonella Typhimurium (79%) and Salmonella Enteritidis (18%). The majority of NTS were isolated during rainy season, and from children ≤ 2 years old. MDR, decreased ciprofloxacin susceptibility, azithromycin and cefotaxime resistance were 80.7%, 4.3%, 3.0% and 2.1% respectively. Resistant rates were higher four antibiotics that is above 80%. Table 9 shows the median resistance rates of Non-typhoid Salmonella to 7 antibiotics.
|
Pathogen |
Antibiotic |
Median of resistance rate% (IQR) |
|
Non-typhoid Salmonella |
Cefotaxime |
2.1 (à) |
|
Nalidixic acid |
4.3 (à) |
|
|
Trimethroprim sulfamethoxazole |
88 (à) |
|
|
Chloramphenicol |
83.7 (à) |
|
|
Azithromycin |
4.3 (à) |
|
|
Ampicillin |
88 (à) |
|
|
Multidrug resistance |
80.7 (à) |
à = IQR could not be calculated because we had less than three papers that assessed resistance rates.
Table 9: Median Resistance rate of Non-typhoid Salmonella to 7 antibiotics in Central Africa, 2008-2018.
3.11 Pseudomonas aeruginosa resistance
Five studies [24, 35, 39, 41, 63] described resistance data from urine, pus and respiratory tract samples, hospital environment; and few invasive (blood) samples. In a study by Cholley et al. [39], resistance rates of clinical isolates of P. aeruginosa to antibiotics remained low in Senegal and Central African Republic. This contrasted with the much higher resistance rate of clinical isolates from Ivory Coast and Nigeria. In this review, at least one study included the use of nosocomial samples from admitted patients. Multi resistance to clinically relevant antibiotics was assessed by Ndip et al. [21] in Cameroon, noted combined resistance to at least two antibiotics in all strains. Table 10 shows the median resistance rates of Pseudomonas spp. to 14 antibiotics.
|
Pathogen |
Antibiotic |
Median of resistance rate% (IQR) |
|
Pseudomonas spp. |
Ceftazidime |
7.1 (à) |
|
Ceftriaxone |
11.4 (à) |
|
|
Ciprofloxacine |
12.7 (à) |
|
|
Gentamycin |
0.5 (à) |
|
|
Chloramphenicol |
41.7 (à) |
|
|
Erythromycin |
34.6 (à) |
|
|
Ampicillin |
58.0 (à) |
|
|
Cotrimoxazole |
75.7 (à) |
|
|
Penicillin |
58.0 (à) |
|
|
Clindamycin |
14.3 (à) |
|
|
Ticarcillin |
25.0 (à) |
|
|
Tobramycin |
7.1 (à) |
|
|
Ampicillinbetalactamaseinhibitor |
81.8 (à) |
|
|
Pipercillintazobactam |
10.7 (à) |
à = IQR could not be calculated because we had less than three papers that assessed resistance rates.
Table 10: Median Resistance rate of Pseudomonas spp. to 14 antibiotics in Central Africa, 2008-2018.
3.12 Klebsiella spp resistance
We obtained data from 8 studies [8, 21, 22, 40-42, 43, 64]. High resistance rates were noted for Ceftazidime (74.9%), Cefuroxime (76.9%), Amoxicillin (91.7%), Amoxillinclav (94.2%), Cotrimoxazole (96.3%), Vancomycin (91.7%), Doxycycline (85.5%), Cotrimoxazole (96.3%). Ndip, et al. [21] (Cameroon) described multiresistance for (combinations of) ampicillin, doxycycline, SXT and chloramphenicol at 77.3% and 33%. From Table 11, it shows the median resistance rates of Klebsiella spp to 23 antibiotics. It is worthy to note that, Klebsiella spp was 100% resistant to 4 of these antibiotics (Trimethroprimsulfamethoxazole, chloramphenicol, Ampicillin and Penicillin).
|
Pathogen |
Antibiotic |
Median of resistance rate% (IQR) |
|
Klebsiella spp |
Ceftazidime |
74.9 (à) |
|
Cefuroxime |
76.9 (à) |
|
|
Ceftriaxone |
47.5 (8-78.8) |
|
|
Cefotaxime |
62.0 (à) |
|
|
Amoxicillin |
91.7 (à) |
|
|
Piperacillin |
12.5 (à) |
|
|
Amoxillinclav |
94.2 (à) |
|
|
Norfloxacin |
46 (à) |
|
|
Ofloxacin |
17 (à) |
|
|
ciprofloxacine |
63.8 (40.4-89.1) |
|
|
gentamycin |
0.8 (0.6-0.9) |
|
|
Vancomycin |
91.7 (à) |
|
|
amikacin |
13.9 (à) |
|
|
Trimethroprimsulfamethoxazole |
100 (à) |
|
|
Chloramphenicol |
100 (à) |
|
|
doxycycline |
85.8 (à) |
|
|
Ampicillin |
100 (100-100) |
|
|
Cotrimoxazole |
96.3 (à) |
|
|
Penicillin |
100 (à) |
|
|
Impenem |
5.8 (à) |
|
|
TMPSMX |
54.0 (à) |
|
|
Cefalothin |
67 (à) |
|
|
Ampicillinbetalactamaseinhibitor |
92.6 (à) |
à = IQR could not be calculated because we had less than three papers that assessed resistance rates.
Table 11: Median Resistance rate of Klebsiella spp. to 23 antibiotics in Central Africa, 2008-2018.
3.13 Escherichia coli resistance
AMR has been reported for E. coli from various animal species, the environment and globally in hospitalized patients. In Central African states, we obtained data from 6 studies [10, 20, 59, 65-67]. Escherichia coli strains that cause diarrhea in humans are either enterotoxigenic E. coli (ETEC), enteroaggregative E. coli (EAEC), enteropathogenic E. coli (EPEC), enteroinvasive E. coli (EIEC), diffusely adherent E. coli (DAEC) or verocytotoxigenic E. coli (VTEC). We retrieved data on E. coli predominantly from urine and faeces, and rarely from blood or pus. The reported and mean resistance rates were high for all commonly used antibiotics but also for amoxicillin/clavulanic acid and first-generation cephalosporins. Table 10 shows the median resistance rates of E. coli to 20 antibiotics.
|
Pathogen |
Antibiotic |
Median of resistance rate% (IQR) |
|
E. Coli |
Ceftazidime |
33 (à) |
|
Cefuroxime |
49.6 (à) |
|
|
Ceftriaxone |
74.1 (à) |
|
|
Cefotaxime |
31 (à) |
|
|
Piperacillin |
8.4 (à) |
|
|
Amoxillinclav |
83.6 (à) |
|
|
Norfloxacin |
39 (à) |
|
|
Ofloxacin |
63 (à) |
|
|
Ciprofloxacine |
53.7 (à) |
|
|
Gentamycin |
.5 (à) |
|
|
Amikacin |
4.9 (à) |
|
|
New_Trimethroprimsulfamethoxazole |
95 (à) |
|
|
chloramphenicol |
66.7 (à) |
|
|
Doxycycline |
78.4 (à) |
|
|
Ampicillin |
96 (à) |
|
|
Cotrimoxazole |
81.2 (à) |
|
|
Penicillin |
81.2 (à) |
|
|
Impenem |
8 (à) |
|
|
TMPSMX |
76 (à) |
|
|
Ampicillinbetalactamaseinhibitor |
62.5 (à) |
à = IQR could not be calculated because we had less than three papers that assessed resistance rates, TMPSMX - Trimethoprim/sulfamethoxazole
Table 12: Median Resistance rate of E. coli to 20 antibiotics in Central Africa, 2008-2018 Enterococci resistance.
There is growing resistance of several bacteria to a class of antibiotics known as cephalosporins and carbapenems, which, for many conditions, represent the “last line” of effective treatment [68]. Carbapenem-resistant Enterobacteriaceae (CRE) are among the most difficult to treat emerging multidrug-resistant organisms. Major limitations exist in surveillance needed to address CRE, particularly in areas with inadequate resources like the countries of Central Africa Some strains of carbapenem resistant. Enterobacteriaceae (CRE) have developed resistance to most of the available antibiotics, resulting in mortality rates of 50% [69, 70].
4. Conclusion
Antimicrobial agents, of which antibiotics are a subset, have saved hundreds of millions of lives from infectious diseases. Antimicrobial resistant (AMR) organisms are increasing globally, threatening to render existing treatments ineffective against many infectious diseases. Drug resistant strains of bacteria, prolong illness, increase case-fatality, facilitate transmission, and increase treatment costs. In summary, our review highlights two important findings: first, there is huge gap of data regarding antimicrobial resistance in the Central African Region. Few studies were found in more than a third of the countries on the region not having recent data on the issue of AMR. Second, a high level of drug resistance exists to commonly prescribed antibiotics in the Central African region. We recommend the standardization and quality of the microbiological identification and susceptibility/resistance testing methods as well as Surveillance to be improved inorder to allow national and international organizations to monitor the extent of the AMR problem. All of the identified areas of concern need urgent attention by the global health community in order to halt the public health threat associated with spreading AMR. In line with Africa CDC [3], initiatives such as the establishment of an Anti-Microbial Resistance Surveillance Network (AMRSNET) is of utmost importance for Africa and particularly in the Central African region.
Declarations
Ethics Approval and Consent to Participate
Not applicable.
Funding
The author (s) received no financial support for the research, authorship, and/or publication of this article.
Consent for Publication
Not applicable.
Availability of Data and Material
Not applicable.
Competing Interests
The authors declare that they have no competing interests.
Acknowledgments
The authors alone are responsible for the views expressed in this article, which do not necessarily represent the views, decisions or policies of the institutions with which they are affiliated.
References
- World Health Organization. United Nations meeting on antimicrobial resistance. WHO (2016).
- World Health Organization, Antimicrobial resistance-SEARO. WHO (2014).
- Africa CDC. Africa CDC Framework for Antimicrobial Resistance, 2018-2023. Africa CDC (2018).
- World Health Organization. Pneumonia WHO (2016).
- World Health Organization. Global Antimicrobial Resistance Surveillance System. WHO (2015).
- Essack SY, Desta AT, Abotsi RE, et al. Antimicrobial resistance in the WHO African region: current status and roadmap for action. J Public Health 39 (2017): 8-13.
- World Health Organization-WHO report on surveillance of antibiotic consumption: 2016-2018 early implementation WHO (2018).
- Kimanga. A Situational Analysis of Antimicrobial Drug Resistance in Africa: Are We Losing the Battle? Ethiop J Health Sci 22 (2012): 135-143.
- Seale AC, Gordon NC, Islam J, et al. AMR Surveillance in low and middle-income settings - A roadmap for participation in the Global Antimicrobial Surveillance System (GLASS). Wellcome Open Res 1 (2017): 2.
- Amin ET, Njumkeng C, Kika BT, et al. Pattern of Antimicrobial Resistance among Bacterial Isolates from Urogenital Clinical Specimens: A Descriptive Study from the Buea Health District, Cameroon. Drugs-Real World Outcomes 5 (2018): 101-108.
- Singh-Babak SD, Babak T, Diezmann S, et al. Global Analysis of the Evolution and Mechanism of Echinocandin Resistance in Candida glabrata. PLOS Pathog 8 (2012): e1002718.
- Debrah I Boeras, Rosanna W Peeling, Philip Onyebujoh, et al. The WHO AFRO external quality assessment programme (EQAP): Linking laboratory networks through EQA programmes 5 (2016): 2.
- Vlieghe E, Phoba MF, Muyembe JJ, et al. Antibiotic resistance among bacterial pathogens in Central Africa: a review of the published literature between 1955 and 2008. HAL Achives-Ouvert 42 (2009).
- Ndihokubwayo JB, Yahaya AA, Abayneh, et al Antimicrobial resistance in the African Region: Issues, challenges and actions proposed. WHO, African Health Monitor (2013).
- Munn Z, Peters MDJ, Stern C, et al. Systematic review or scoping review? Guidance for authors when choosing between a systematic or scoping review approach. BMC Med Res Methodol 19 (2018): 18.
- Impellizzeri FM, Bizzini M. Systematic Review and Meta?Analysis: A Primer. Int J Sports Phys Ther 7 (2012): 493-503.
- Booth A, Moore G, Flemming K, et al. Taking account of context in systematic reviews and guidelines considering a complexity perspective. BMJ Glob Health 4 (2019): e000840.
- Pham MT, Raji? A, Greig JD, et al. A scoping review of scoping reviews: advancing the approach and enhancing the consistency. Res Synth Methods 5 (2014): 371-385.
- Minime-Lingoupou F, Manirakiza A, Yango F, et al. Relatively low primary resistance to anti-tuberculosis drugs in Bangui and Bimbo, Central African Republic. Int J Tuberc Lung J Int Union Tuberc Lung Dis 5 (2011): 657-661.
- Akoachere JFT, Yvonne S, Akum NH, et al. Etiologic profile and antimicrobial susceptibility of community-acquired urinary tract infection in two Cameroonian towns. BMC Res Notes 5 (2012): 219.
- Ndip RN, Ntiege EA, Ndip LM, et al. Antimicrobial Resistance of Bacterial Agents of the Upper Respiratory Tract of School Children in Buea, Cameroon. J Health POPUL NUTR 26 (2008): 8.
- Nzalie RN, Gonsu HK, Koulla-Shiro S. Bacterial Etiology and Antibiotic Resistance Profile of Community-Acquired Urinary Tract Infections in a Cameroonian City. International Journal of Microbiology (2016).
- Njunda AL, Assob JC, Nsagha DS, et al. Epidemiological, clinical features and susceptibility pattern of shigellosis in the Buea health district, Cameroon. BMC Res Notes 5 (2012): 54.
- Gonsu KH, Ndongo GA, Adiogo D, et al. Carriage of multi-drug resistant bacteria among medical staff of the Yaoundé University Teaching Hospital, Cameroon. J Bacteriol Res 5 (2013).
- Phoba M-F, Lunguya O, Mayimon DV, et al. Multidrug-Resistant Salmonella enterica, Democratic Republic of the Congo. Emerg Infect Dis 18 (2012): 1692-1694.
- Lunguya O, Lejon V, Phoba M-F, et al. Salmonella Typhi in the Democratic Republic of the Congo: Fluoroquinolone Decreased Susceptibility on the Rise. PLoS Negl Trop Dis 6 (2012).
- Kalonji LM, Post A, Phoba M-F, et al. Invasive Salmonella Infections at Multiple Surveillance Sites in the Democratic Republic of the Congo, 2011–2014. Clin Infect Dis 61 (2015): 346-353.
- Lunguya O, Lejon V, Phoba MF, et al. Antimicrobial Resistance in Invasive Non-typhoid Salmonella from the Democratic Republic of the Congo: Emergence of Decreased Fluoroquinolone Susceptibility and Extended-spectrum Beta Lactamases. PLoS Negl Trop Dis 7 (2013): e2103.
- Falay D, Kuijpers LMF, Phoba M-F, et al. Microbiological, clinical and molecular findings of non-typhoidal Salmonella bloodstream infections associated with malaria, Oriental Province, Democratic Republic of the Congo. BMC Infect Dis16 (2016): 271.
- Smith AM, Njanpop-Lafourcade BM, Mengel MA, et al. Comparative Characterization of Vibrio cholerae O1 from Five Sub-Saharan African Countries Using Various Phenotypic and Genotypic Techniques. PLOS ONE 10 (2015): e0142989.
- Schaumburg F, Ngoa UA, Kosters K, et al. Virulence factors and genotypes of Staphylococcus aureus from infection and carriage in Gabon. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis 17 (2011): 1507-1513.
- Peterson AE, Mechan F, Julian KG, et al. Molecular and phenotypic characteristics of healthcare- and community-associated methicillin-resistant Staphylococcus aureus at a rural hospital. PloS One 7 (2012): e38354.
- Schaumburg F, Alabi AS, Mombo-Ngoma G, et al. Transmission of Staphylococcus aureus between mothers and infants in an African setting. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis 20 (2014): 390-396.
- Schaumburg F, Alabi AS, Kaba H, et al. Molecular characterization of Shigella spp. from patients in Gabon 2011-2013. Trans R Soc Trop Med Hyg 109 (2015): 275-279.
- Alabi AS, Frielinghaus L, Kaba H, et al. Retrospective analysis of antimicrobial resistance and bacterial spectrum of infection in Gabon, Central Africa. BMC Infect Dis 13 (2013): 455.
- Bercion R, Njuimo SP, Boudjeka PM, et al. Distribution and antibiotic susceptibility of Shigella isolates in Bangui, Central African Republic. Trop Med Int Health 13 (2008): 468-471.
- Yaya EL, Djeintote M, Djimeli CL, et al. Contribution to the Study of Antibiotic Resistance on Salmonella and Shigella Strains Isolated in Central African Republic. J Microbiol Exp 4 (2017): 1.
- Mossoro-Kpinde CD, Manirakiza A, Mbecko JR, et al. Antimicrobial Resistance of Enteric Salmonella in Bangui, Central African Republic. J Trop Med (2015): 2-5.
- Cholley P, Ka R, Guyeux C, et al. Population Structure of Clinical Pseudomonas aeruginosa from West and Central African Countries. PLOS ONE 9 (2014): e107008.
- Makeda AC. Five year resistance Trends of Bacterial isolates in Kigali, Rwanda. Yale EliScholar (2015).
- Carroll M, Rangaiahagari A, Musabeyezu E, et al. Five-Year Antimicrobial Susceptibility Trends among Bacterial Isolates from a Tertiary Health-Care Facility in Kigali, Rwanda. Am J Trop Med Hyg 95 (2016): 1277.
- Umuhoza C, Barton T. 983Outbreak of multi-drug resistant Klebsiella pneumoniae: A 4 month epidemiologic follow-up in a tertiary teaching hospital in Rwanda. Open Forum Infect Dis 1 (2014): 286.
- Ntirenganya C, Manzi O, Muvunyi CM, et al. High Prevalence of Antimicrobial Resistance Among Common Bacterial Isolates in a Tertiary Healthcare Facility in Rwanda. Am J Trop Med Hyg 92 (2015): 865-870.
- Abdelhadi O, Ndokaïn J, Ali MM, et al. [Drug resistance testing of Mycobacterium tuberculosis isolates from sputum in Chad]. Bull Soc Pathol Exot 1990 105 (2012): 16-22.
- Ekouya Bowassa G, Ontsira-Ngoyi EN, Okoko AR, et al. Bacteriology of early neonatal infection in Brazzaville (Congo). Arch Pediatr Organe Off Soc Francaise Pediatr 22 (2015): 1099-1101.
- Shatalov A. Prevalence and Antibiotic Resistance Pattern of Escherichia coli and Klebsiella pneumoniae in Urine Tract Infections at the La Paz Medical Center, Malabo, Equatorial Guinea. Open J Med Microbiol 05 (2015): 177.
- Conceicao T, Santos Silva I, de Lencastre H, et al. Staphylococcus aureus nasal carriage among patients and health care workers in São Tomé and Príncipe. Microb Drug Resist Larchmt N 20 (2014): 57-66.
- Noeske J, Foe JL, Kuaban C. Cameroon’s MDR-TB treatment programme jeopardised by cross-border migration. Eur Respir J 47 (2016): 684-686.
- Gavín P, Iglesias MJ, Jiménez MS, et al. Multidrug-Resistant Mycobacterium tuberculosis Strain from Equatorial Guinea Detected in Spain. Emerg Infect Dis 15 (2009): 1858-1860.
- Umubyeyi AN, Vandebriel G, Gasana M, et al. Results of a national survey on drug resistance among pulmonary tuberculosis patients in Rwanda. Int J Tuberc Lung Dis Off J Int Union Tuberc Lung Dis 11 (2007): 189-194.
- Akoachere JFTK, Masalla TN, Njom HA. Multi-drug resistant toxigenic Vibrio cholerae O1 is persistent in water sources in New Bell-Douala, Cameroon. BMC Infect Dis 13 (2013): 366.
- Bercion R, Bobossi-Serengbe G, Gody JC, et al. Acute bacterial meningitis at the “Complexe Pédiatrique” of Bangui, Central African Republic. J Trop Pediatr 54 (2008): 125-128.
- Falagas, Karageorgopoulos D, Leptidis J, Korbila P. MRSA in Africa: Filling the Global Map of Antimicrobial Resistance. PLOS ONE 8 (2013): e68024.
- Straub L, Stegger M, Akpaka PE, et al. Origin, evolution, and global transmission of community-acquired Staphylococcus aureus ST8. Proc Natl Acad Sci 114 (2017): 10596-10604.
- Abdulgader SM, Shittu AO, Nicol MP, et al. Molecular epidemiology of Methicillin-resistant Staphylococcus aureus in Africa: a systematic review. Front Microbiol 1 (2015): 6.
- Shittu AO, Kaba M, Abdulgader MS, et al. Mupirocin-resistant Staphylococcus aureus in Africa: a systematic review and meta-analysis. Antimicrob Resist Infect Control (2018): 7.
- Breurec S, Rafaï C, Onambele M, et al. Serotype Distribution and Antimicrobial Resistance of Shigella Species in Bangui, Central African Republic, from 2002 to 2013. Am J Trop Med Hyg 99 (2018): 283-286.
- Akoachere JFT, Tanih N, Ndip LM, et al. Phenotypic Characterization of Salmonella Typhimurium Isolates from Food-animals and Abattoir Drains in Buea, Cameroon. J Health Popul Nutr 27 (2009): 612-618.
- Tadesse BT, Ashley EA, Ongarello S, et al. Antimicrobial resistance in Africa: a systematic review. BMC Infect Dis (2017): 17.
- Tadesse G, Tessema TS, Beyene G, et al. Molecular epidemiology of fluoroquinolone resistant Salmonella in Africa: A systematic review and meta-analysis. PLOS ONE 13 (2018): e0192575.
- Phoba MF, Barbé B, Lunguya O, et al. Salmonella enterica serovar Typhi Producing CTX-M-15 Extended Spectrum β-Lactamase in the Democratic Republic of the Congo. Clin Infect Dis Off Publ Infect Dis Soc Am 65 (2017): 1229-1231.
- Antunes P, Campos J, Mourao J, et al. High occurrence and unusual serotype diversity of non-typhoidal Salmonella in non-clinical niches, Angola. Epidemiol Infect 145 (2017): 883-886.
- Rickard J, Ngarambe C, Ndayizeye L, et al. Antibiotic Use and Antimicrobial Resistance of Surgical Patients with Peritonitis at a Tertiary Referral Hospital in Rwanda. Surg Infect 19 (2018): 382-387.
- Shatalov A. Prevalence and Antibiotic Resistance Pattern of Escherichia coli and Klebsiella pneumoniae in Urine Tract Infections at the La Paz Medical Center, Malabo, Equatorial Guinea. Open J Med Microbiol 5 (2015): 177.
- Stoesser N, Sheppard AE, Pankhurst L, et al. Evolutionary History of the Global Emergence of the Escherichia coli Epidemic Clone ST131. mBio 7 (2016).
- Inge DJ, Levine MM, Kotloff KL, et al. Dynamics of antimicrobial resistance in intestinal Escherichia coli from children in community settings in South Asia and sub-Saharan Africa. Nat Microbiol 3 (2018): 1063.
- Lupindu AM. Epidemiology of Shiga toxin-producing Escherichia coli O157:H7 in Africa in review. South Afr J Infect Dis 33 (2018): 24-30.
- Ventola CL. The Antibiotic Resistance Crisis: Part 1: Causes and Threats. Pharm Ther 40 (2015): 277
- CDC. Carbapenem-resistant Enterobacteriaceae in Healthcare Settings, HAI,| CDC (2018).
- Chiotos K Han, Tamma PD. Carbapenem-Resistant Enterobacteriaceae Infections in Children. Curr Infect Dis Rep 18 (2016): 2.
|
Citation: Patrick Achiangia Njukeng, Denis Ebot Ako-Arrey, Elvis Tajoache Amin, Charles Njumkeng, Frankline Sevidzem Wirsiy. Antimicrobial Resistance in the Central African Region: A Review. Journal of Environmental Science and Public Health 3 (2019): 358-378. |
